最近对16s的三大数据库的序列的具体序列情况挺好奇的,决定统计一下各个序列的长度分布情况,以及这些序列具体分布在哪几个V区,有助于我解决后面16So数据的问题。还是用上我三脚猫的功夫,开始今天 的探索,没有人探索的事情,还是挺开心的。
1.统计序列长度分布情况
#获得长度列表文件
length_list = []
with open('current_Bacteria_unaligned.fa') as f:
flag = 0
length = 0
for line in f:
if line.startswith('>'):
flag = 1
if length != 0:
#print(length)
length_list.append(length)
#break
length = 0
else:
length = 0
elif flag == 1:
length +=len(line)
fout = open('length_table.txt', 'w')
for a in length_list:
fout.write(str(a) + '\n')
fout.close()
#统计长度区间分布
length_150 = 0
length_300 = 0
length_600 = 0
length_1300 = 0
length_1300_ = 0
with open('length_table.txt') as f:
for line in f:
if 0 < int(line.strip()) <= 150:
length_150 += 1
elif 150 < int(line.strip()) <= 300:
length_300 += 1
elif 300 < int(line.strip()) <= 600:
length_600 += 1
elif 600 < int(line.strip()) <= 1300:
length_1300 += 1
elif int(line.strip()) > 1300:
length_1300_ += 1
print('150以内:', length_150,'\n',
'150-300:', length_300, '\n',
'300-600:', length_600, '\n',
'600-1300:', length_1300,'\n',
'1300-:', length_1300_)
#最后结果 150以内: 0 150-300: 0 300-600: 617,554 600-1300: 1,063,377 1300-: 1,515,109
我也可视化一下:
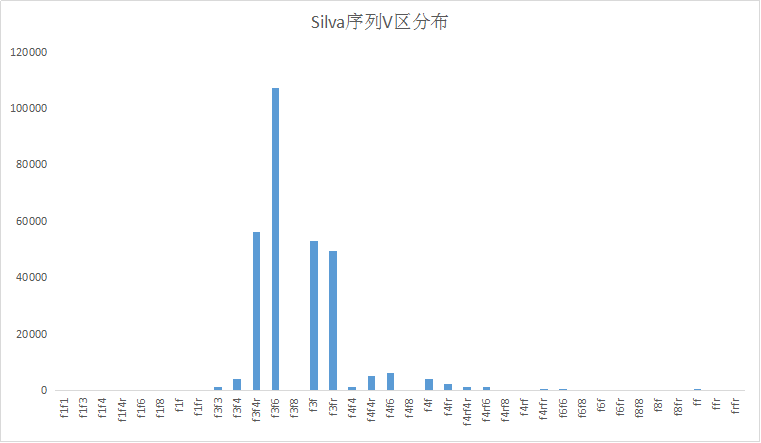
更正一下,这里用的是RDP数据库,之前由于Silva数据库用的是不兼容的14级分类系统而没采用。
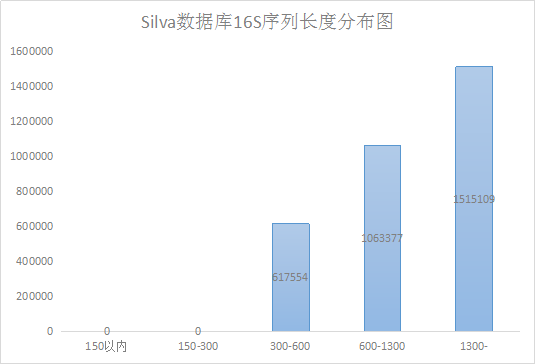
从图中可以看出,大部分序列还是集中在600以上。
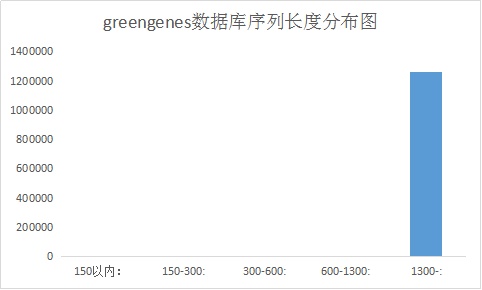
接着是greengenes数据库,这个数据库虽然序列较少,但是长度大部分集中在1300+,质量较高,就是好久没更新过了。
150以内: 0
150-300: 0
300-600: 0
600-1300: 895
1300-: 1262090
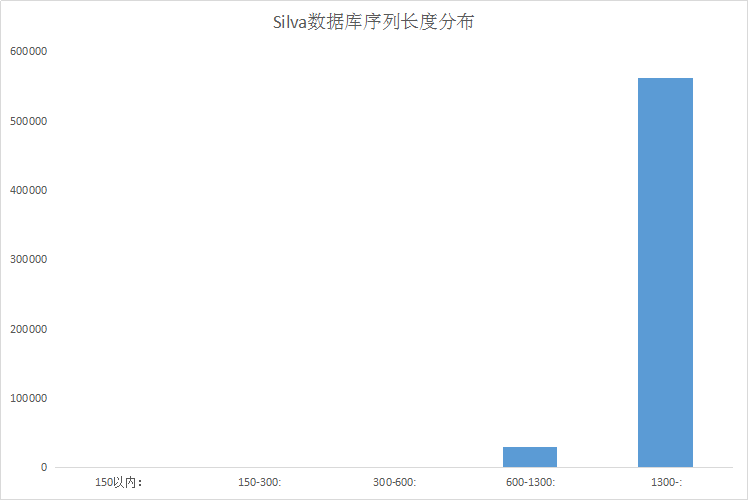
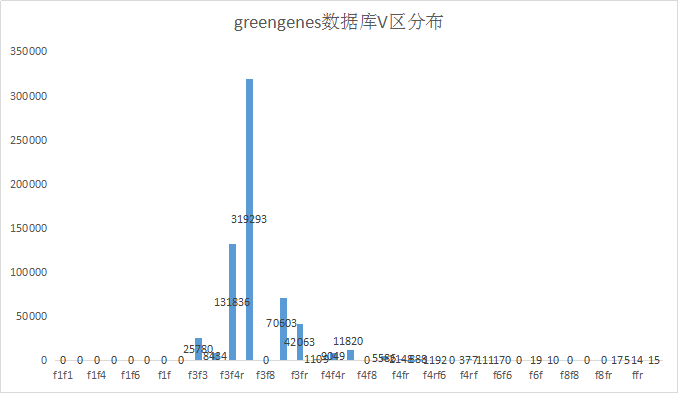
2.统计V区分布情况
从一个公众号得到的一张分布图是这样的,
![]()
![]()
![]()
![]()
我想确定的是序列都包含在哪个或者哪两个区。花了好大有力气,才把代码理好,效率应该还是比较低的,但是,对于我来说
import re
dic ={}
v1f = re.compile('GGATCCAGACTTTGATYMTGGCTCAG', re.I)
v3f = re.compile('CCTA[CT]GGG[AG][GTC]GCA[CG]CAG', re.I)
v4f = re.compile('GTG[CT]CAGC[AC]GCCGCGGTAA', re.I)
v4r = re.compile('ATTAGA[AT]ACCC[CTG][ATGC]GTAGTCC', re.I)
v6f = re.compile('AACGCGAAGAACCTTAC', re.I)
v8f = re.compile('CGTCATCC[AC]CACCTTCCTC', re.I)
vr = re.compile('AAGTCGTAACAAGGTA[AG]CCGTA', re.I)
#v4r = re.compile('GGACTAC[ATGC][ACG]GGGT[AT]TCTAAT', re.I)
vf = re.compile('AG[AG]GTT[CT]GAT[CT][AC]TGGCTCAG', re.I)
#vr = re.compile('GACGGGCGGTG[AT]GT[AG]CA', re.I)
#seq_list = []
def decide_which_zone(f1, f3, f4, f4r, f6, f8, f, fr):
type = [f1, f3, f4, f4r, f6, f8, f, fr]
type_str = ['f1', 'f3', 'f4', 'f4r', 'f6', 'f8', 'f', 'fr']
type_name = []
for i in range(len(type)):
#print(type[i])
if type[i] != None :
type_name.append(type_str[i])
else:
continue
if i + 1 < len(type):
if type[i + 1] != None and type_str[i + 1] not in type_str:
type_name.append(type_str[i + 1])
else:
continue
if i + 2 < len(type):
if type[i + 2] != None and type_str[i + 2] not in type_str:
type_name.append(type_str[i + 2])
else:
continue
if i + 3 < len(type):
if type[i + 3] != None and type_str[i + 3] not in type_str:
type_name.append(type_str[i + 3])
else:
continue
if i + 4 < len(type):
if type[i + 4] != None and type_str[i + 4] not in type_str:
type_name.append(type_str[i + 4])
else:
continue
if i + 5 < len(type):
if type[i + 5] != None and type_str[i +5 ] not in type_str:
type_name.append(type_str[i + 5])
else:
continue
if i + 6 < len(type):
if type[i + 6] != None and type_str[i + 6] not in type_str:
type_name.append(type_str[i + 6])
else:
continue
if i + 7 < len(type):
if type[i + 7] != None and type_str[i + 7] not in type_str:
type_name.append(type_str[i + 7])
else:
continue
return type_name
with open('current_Bacteria_unaligned.fa') as f:
li = ['f1', 'f3', 'f4', 'f4r', 'f6', 'f8', 'f', 'fr']
for i in range(len(li)):
for j in range(len(li)):
if i <= j:
dic[li[i] + li[j]] = 0
#print(dic)
flag = 0
seq = ''
#i = 0
for line in f:
if line.startswith('>'):# and i <=2:
flag = 1
if seq != '':
#print(seq)
f1 = v1f.search(seq)
f4r = v4r.search(seq)
f3 = v3f.search(seq)
f4 = v4f.search(seq)
f6 = v6f.search(seq)
f8 = v8f.search(seq)
f = vf.search(seq)
fr = vr.search(seq)
#seq_list.append(length)
type_name = []
type_name = decide_which_zone(f1, f3, f4, f4r, f6, f8, f, fr)
print(type_name)
if type_name != []:
type_key = type_name[0] + type_name[-1]
dic[type_key] += 1
#i += 1
#break
seq = ''
flag = 0
else:
seq = ''
elif flag == 1:
seq += line.strip()
print(dic)
#运行结果
{'f1f1': 0, 'f1f3': 0, 'f1f4': 1, 'f1f4r': 0, 'f1f6': 0, 'f1f8': 0, 'f1f': 0, 'f1fr': 0, 'f3f3': 120867, 'f3f4': 143686, 'f3f4r': 356027, 'f3f6': 400108, 'f3f8': 4, 'f3f': 171637, 'f3fr': 53716, 'f4f4': 17211, 'f4f4r': 71719, 'f4f6': 40223, 'f4f8': 4, 'f4f': 13185, 'f4fr': 3646, 'f4rf4r': 39537, 'f4rf6': 45160, 'f4rf8': 0, 'f4rf': 977, 'f4rfr': 4263, 'f6f6': 43983, 'f6f8': 0, 'f6f': 64, 'f6fr': 2300, 'f8f8': 0, 'f8f': 0, 'f8fr': 0, 'ff': 1670, 'ffr': 26, 'frfr': 3678}
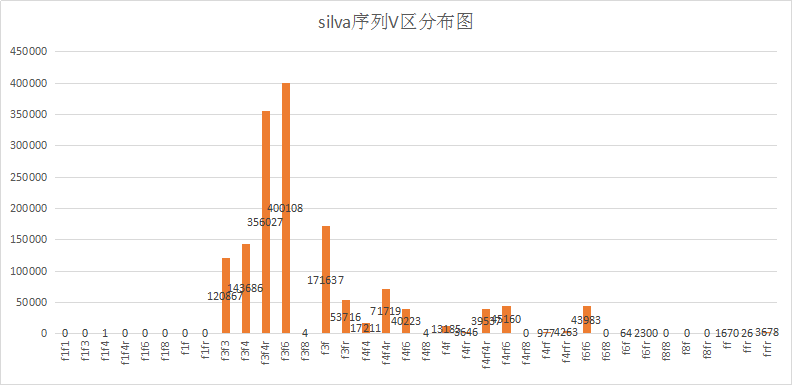
上图依然是RDP数据库,前面标错了。应该是某个地方出了问题,序列不全,少了估计有一半。不过趋势还是比较明显的,V3-V4是最多的,这也是前些年主流的研究的测序方式。