如果你不懂代码,不懂网站规则,那么最简单的就是直接使用UCSC xena 浏览器啦!!!网站;https://xenabrowser.net/datapages/ 理论上也可以完成大部分数据探索的,甚至还有一些sci文章发表就是完完全全使用这个网页工具。
看到jimmy总结的如此有规律的下载地址链接,我尝试用python写几句脚本下载一下tcga数据。
1.尝试用爬虫获得所有疾病条目
尝试写爬虫发现网页需要javascript,暂时没有搞定,于是偷个懒把内容从https://xenabrowser.net/datapages/?host=https://tcga.xenahubs.net复制下来保存为TCGA_names.txt。一共是39 Cohorts, 903 Datasets。
TCGA Acute Myeloid Leukemia (LAML)
TCGA Adrenocortical Cancer (ACC)
……
- 把这些文本解析一下,变成下载地址
https://gdc.xenahubs.net/download/TCGA-LAML/Xena_Matrices/TCGA-LAML.htseq_counts.tsv.gz
https://gdc.xenahubs.net/download/TCGA-ACC/Xena_Matrices/TCGA-ACC.htseq_counts.tsv.gz
绝大部分文件的下载地址中,网址的规律是:
https://gdc.xenahubs.net/download/TCGA-疾病名称/Xena_Matrices/TCGA-疾病名称-下载文件.tsv.gz
基本思路就是定义了两个函数完成这个事。第一个函数 get_name() 是用来从文本中获取疾病名的缩写,也就是这三十几种,因为下载地址就是用的缩写,利用 split() 函数获得。
import os
def get_name(file_in):
name_list= []
for line in file_in:
if ‘TCGA’ in line:
name_list.append(line.strip().split(‘(‘)\ [1].split(‘)’)[0])
return name_list
第二个函数是把下载地址补充完整,实现下载过程。首先看你需要的数据是哪几类,把网址里的文件名放在一个列表里。这里数据文件名和含义的对应关系是:
gene expression RNAseq(HTSeq)–htseq_counts
survival–survival
phenotype–GDC_phenotype
miRNA Expression Quantification–mirna
copy number–masked_cnv
MuSE Variant Aggregation and Masking–muse_snv
MuTect2 Variant Aggregation and Masking–mutect2_snv
然后是利用格式化字符串,实现网址的自动补全。最后,利用os.system()函数,调用wget命令完成,-c参数以实现断点续传。
def download_files(name_list):
for name in name_list:
file_list = [‘htseq_counts’, ‘survival’,’GDC_phenotype’, ‘mirna’, ‘masked_cnv’,’muse_snv’, ‘mutect2_snv’]
for i in range(len(file_list)):
cmd = ‘wget -c https://gdc.xenahubs.net/download/TCGA-%s/Xena_Matrices/TCGA-%s.%s.tsv.gz’ % (name, name, file_list[i])
os.system(cmd)
最后,调用两个函数,完成下载过程。
file_in = open(‘TCGA_names.txt’)
name_list = get_name(file_in)
download_files(name_list)
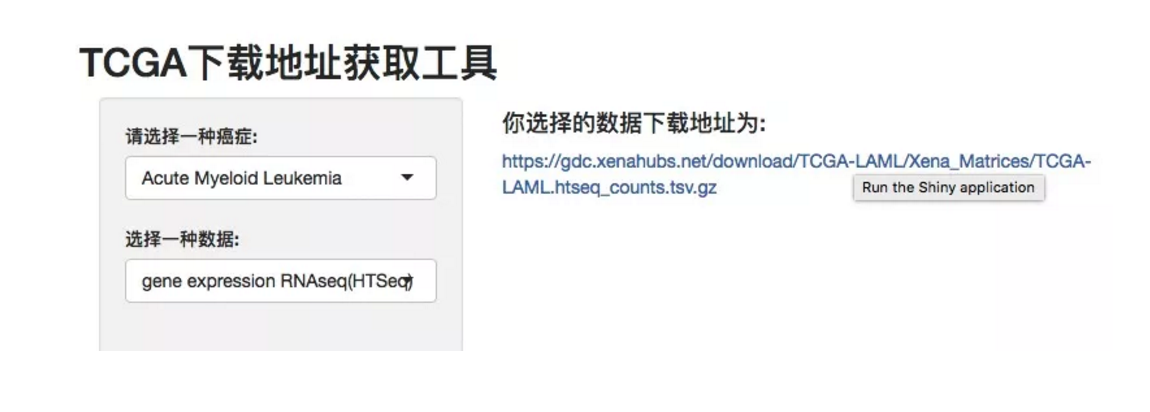
3.做个简陋的shiny控件
在循循善诱的群主启发下第一次学着用shiny,有点赶鸭子上架,哈哈。就参考着shiny的官方教程,做了个能有多简单就多简单的。
UI代码,就是按照官方示例框架,加上了内容:
library(shiny)
Define UI for miles per gallon application
shinyUI(list(
Application title
#titlePanel(“TCGA下载地址获取工具”),
fluidPage(
titlePanel(“TCGA下载地址获取工具”),
sidebarPanel(
selectInput(“variable”, “请选择一种癌症:”,
list(“Acute Myeloid Leukemia” = “LAML”,
“Adrenocortical Cancer” = “ACC”,
“Bile Duct Cancer” = “CHOL”,
“Bladder Cancer” = “BLCA”,
“Breast Cancer” = “BRCA”,
“Cervical Cancer” = “CESC”,
“Colon and Rectal Cancer” = “COADREAD”,
“Colon Cancer” = “COAD”,
“Endometrioid Cancer” = “UCEC”,
“Esophageal Cancer” = “ESCA”,
“Formalin Fixed Paraffin-Embedded Pilot Phase II” = “FPPP”,
“Glioblastoma” = “GBM”,
“Head and Neck Cancer” = “HNSC”,
“Kidney Chromophobe” = “KICH”,
“Kidney Clear Cell Carcinoma” = “KIRC”,
“Kidney Papillary Cell Carcinoma” = “KIRP”,
“Large B-cell Lymphoma” = “DLBC”,
“Liver Cancer” = “LIHC”,
“Lower Grade Glioma” = “LGG”,
“lower grade glioma and glioblastoma” = “GBMLGG”,
“Lung Adenocarcinoma” = “LUAD”,
“Lung Cancer” = “LUNG”,
“Lung Squamous Cell Carcinoma” = “LUSC”,
“Melanoma” = “SKCM”,
“Mesothelioma” = “MESO”,
“Ocular melanomas” = “UVM”,
“Ovarian Cancer” = “OV”,
“Pan-Cancer” = “PANCAN”,
“PANCAN12” = “PANCAN12”,
“Pancreatic Cancer” = “PAAD”,
“Pheochromocytoma & Paraganglioma” = “PCPG”,
“Prostate Cancer” = “PRAD”,
“Rectal Cancer” = “READ”,
“Sarcoma” = “SARC”,
“Stomach Cancer” = “STAD”,
“Testicular Cancer” = “TGCT”,
“Thymoma” = “THYM”,
“Thyroid Cancer” = “THCA”,
“Uterine Carcinosarcoma” = “UCS”
)),
selectInput(“var”, “选择一种数据:”,
list(“gene expression RNAseq(HTSeq)” = “htseq_counts”,
“survival” = “survival”,
“phenotype” = “GDC_phenotype”,
“miRNA Expression Quantification” = “mirna”,
“copy number” = “masked_cnv”,
“MuSE Variant Aggregation and Masking” = “muse_snv”,
“MuTect2 Variant Aggregation and Masking” = “mutect2_snv”))
#checkboxInput("outliers", "Show outliers", FALSE)
),
mainPanel(h4("你选择的数据下载地址为:"),
htmlOutput("mySite"))
)))
Server端代码就是同样把地址拼接了一下,同样是简单修改自模板,开始空格问题没搞定,后来百度了个解决方案,但是提示语却也不能有空格了,只好用下划线代替:
library(shiny)
Define server logic required to plot various variables against mpg
shinyServer(
function(input, output, session)
{
output$mySite <- renderUI( {
url <- paste(‘\n’, (gsub(“([N ])”, “”, paste(“https://gdc.xenahubs.net/download/TCGA-“, input$variable,”/Xena_Matrices/TCGA-“,input$variable,”.”,input$var,”.tsv.gz”))))
tags$a(href = url, url)
})
}
)
最后来看看简陋的效果: