最近开始阅读一本关于机器学习的新书,分享一些笔记给大家!
挖掘RMS Titanic数据集
六个阶段
提出问题、数据采集、数据清洗、基础数据分析、高级分析和模型评估 直接上代码呀!数据下载,需要科学地上网下载地址
# 数据下载
train.data <- read.csv("train.csv", na.strings = c("NA", ""))
# 类型转换,分类变量转因子
train.data$Survived <- factor(train.data$Survived)
train.data$Pclass <- factor(train.data$Pclass)
# 检测缺失
sum(is.na(train.data$Age) == TRUE)/length(train.data$Age)
sapply(train.data, function(df){
sum(is.na(df)==TRUE)/length(df)
})
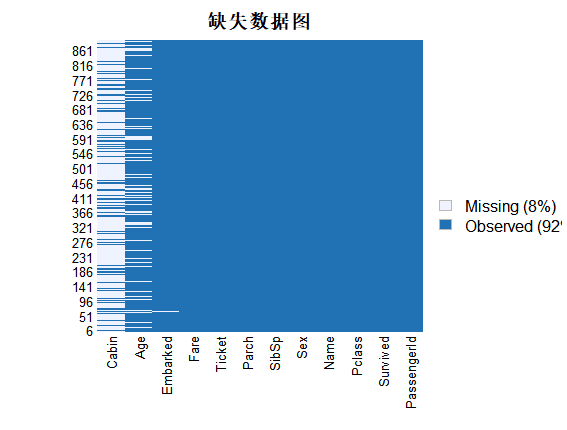
# 缺失数据可视化
# install.packages("Amelia")
require(Amelia)
missmap(train.data, main = "缺失数据图")
AmeliaView()
 AmeliaView有交互式的GUI,赞一个!
AmeliaView有交互式的GUI,赞一个!
table(train.data$Embarked,useNA = "always")
# C Q S <NA>
168 77 644 2
# 将缺失处理为理可能结果
train.data$Embarked[which(is.na(train.data$Embarked))] <- 'S'
table(train.data$Embarked,useNA = "always")
# C Q S <NA>
168 77 646 0
# 获得不同称呼类别
train.data$Name <- as.character(train.data$Name)
# 先用空白标记
table_words <- table(unlist(strsplit(train.data$Name, "\s+")))
str(table_words)
# 'table' int [1:1673(1d)] 1 1 1 1 1 1 1 1 1 1 ...
# - attr(*, "dimnames")=List of 1
# ..$ : chr [1:1673] ""Andy"" ""Annie" ""Annie"" ""Archie"" ...
sort(table_words [grep('\.',names(table_words))],
decreasing = TRUE)
# Mr. Miss. Mrs. Master. Dr. Rev.
517 182 125 40 7 6
Col. Major. Mlle. Capt. Countess. Don.
2 2 2 1 1 1
Jonkheer. L. Lady. Mme. Ms. Sir.
1 1 1 1 1 1
识别和可视化
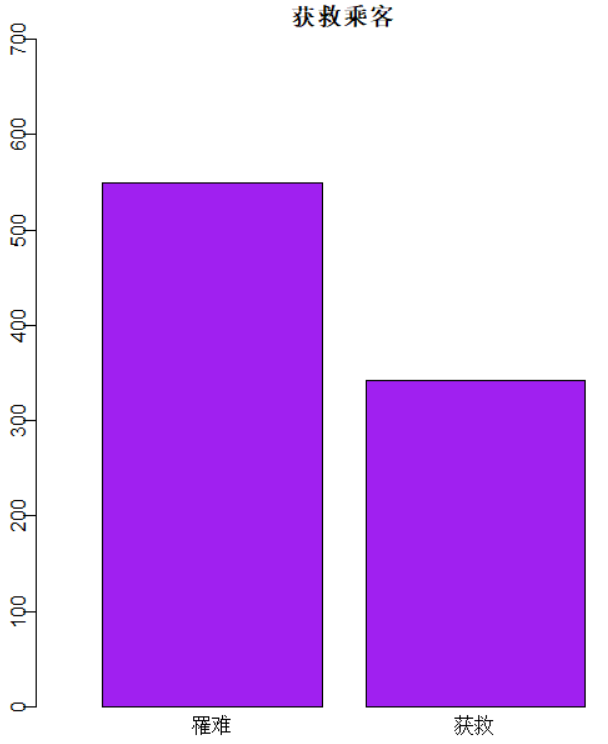

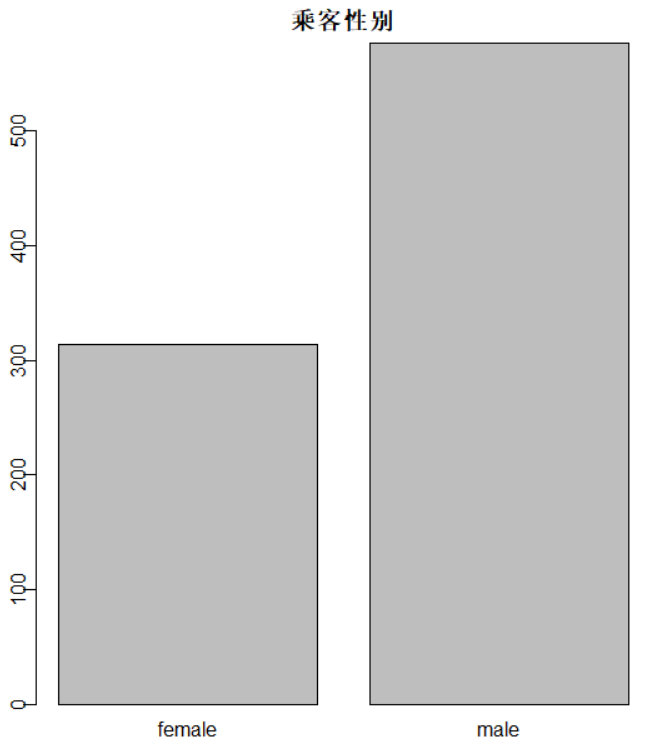




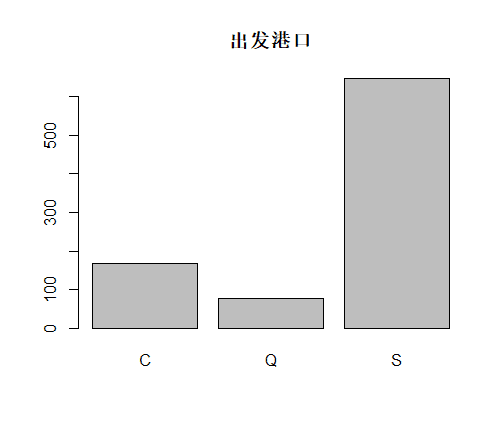
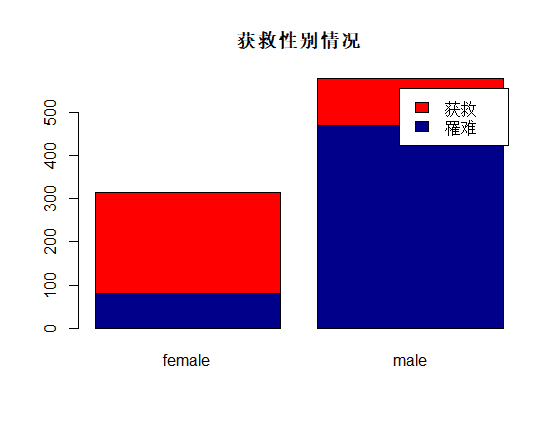
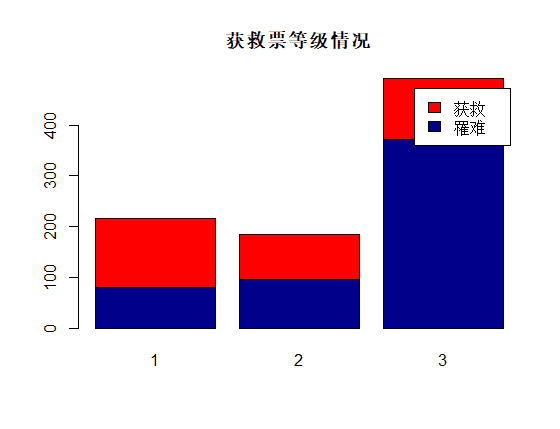
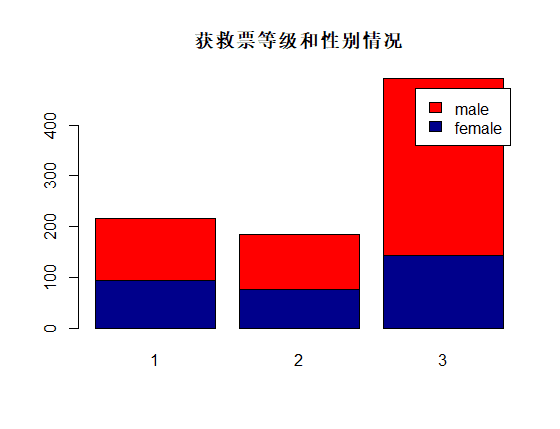
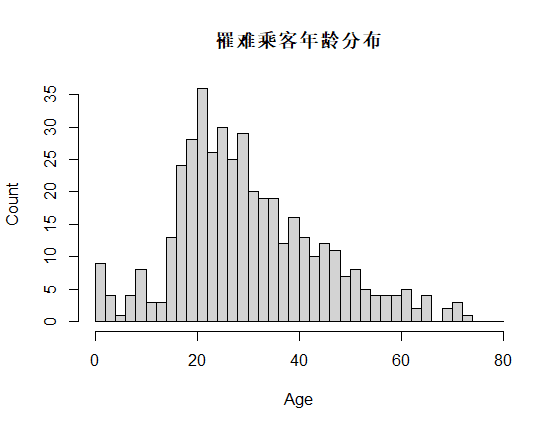
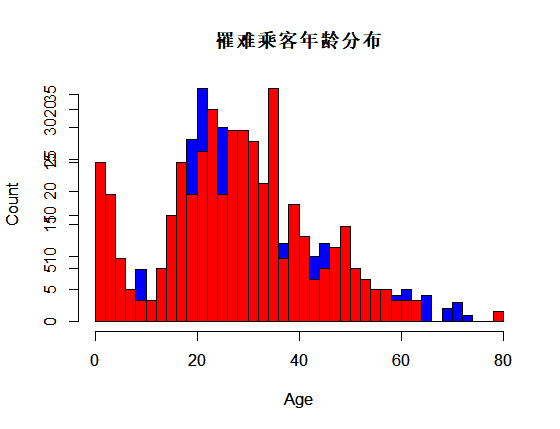
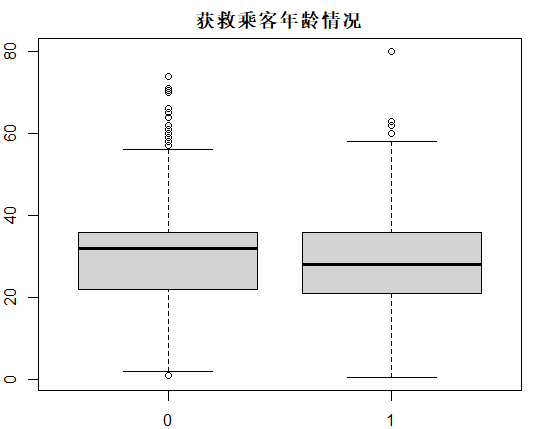
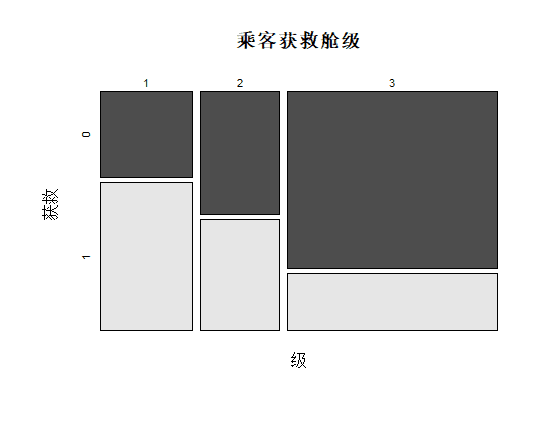
决策树构建
Conditional inference tree with 6 terminal nodes
Response: Survived
Inputs: Pclass, Sex, Age, SibSp, Fare, Parch, Embarked
Number of observations: 623
1) Sex == {female}; criterion = 1, statistic = 180.207
2) Pclass == {3}; criterion = 1, statistic = 59.081
3)* weights = 92
2) Pclass == {1, 2}
4)* weights = 111
1) Sex == {male}
5) Pclass == {1}; criterion = 1, statistic = 21.732
6) Age <= 38; criterion = 0.99, statistic = 10.105
7)* weights = 57
6) Age > 38
8)* weights = 39
5) Pclass == {2, 3}
9) Age <= 3; criterion = 1, statistic = 22.354
10)* weights = 12
9) Age > 3
11)* weights = 312
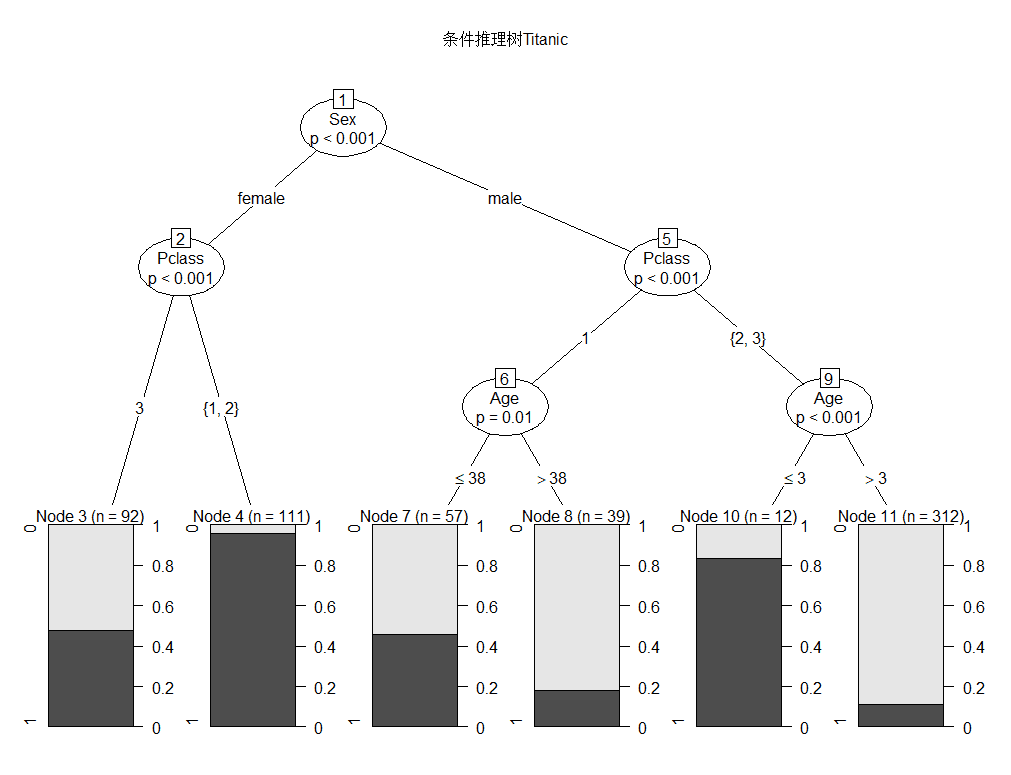
party包的决策树,与rpart包的相比,可以避免rpart包在变量选择时的倾斜,并且更倾向于选择能够产生更多分支或缺失值多的变量。
Confusion Matrix and Statistics
Reference
Prediction 0 1
0 146 57
1 7 57
Accuracy : 0.7603
95% CI : (0.7045, 0.8102)
No Information Rate : 0.573
P-Value [Acc > NIR] : 1.279e-10
Kappa : 0.4811
Mcnemar's Test P-Value : 9.068e-10
Sensitivity : 0.9542
Specificity : 0.5000
Pos Pred Value : 0.7192
Neg Pred Value : 0.8906
Prevalence : 0.5730
Detection Rate : 0.5468
Detection Prevalence : 0.7603
Balanced Accuracy : 0.7271
'Positive' Class : 0

本篇文章来源于微信公众号: 微因